Wastewater-based SARS-CoV-2 epidemiology (WBE) has proven as an excellent tool to monitor pandemic dynamics supporting individual testing strategies. WBE can also be used as an early warning system for monitoring the emergence of novel pathogens or viral variants. However, for a timely transmission of results, sophisticated sample logistics and analytics performed in decentralized laboratories close to the sampling sites are required. Since multiple decentralized laboratories commonly use custom in-house workflows for sample purification and PCR-analysis, comparative quality control of the analytical procedures is essential to report reliable and com-
parable results.
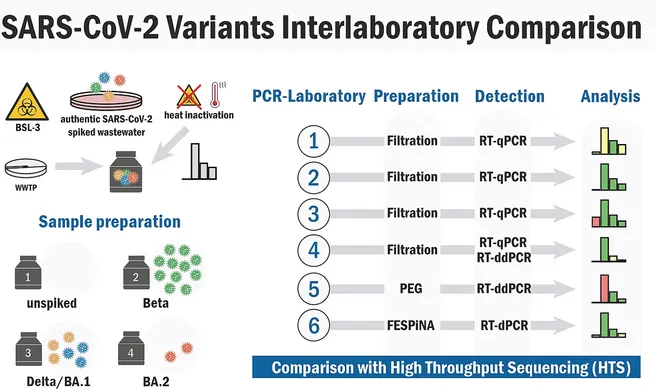
In this study, we performed an interlaboratory comparison at laboratories specialized for PCR and high- throughput-sequencing (HTS)-based WBE analysis. Frozen reserve samples from low COVID-19 incidence pe- riods were spiked with different inactivated authentic SARS-CoV-2 variants in graduated concentrations and ratios. Samples were sent to the participating laboratories for analysis using laboratory specific methods and the reported viral genome copy numbers and the detection of viral variants were compared with the expected values.
All PCR-laboratories reported SARS-CoV-2 genome copy equivalents (GCE) for all spiked samples with a mean intra- and inter-laboratory variability of 19 % and 104 %, respectively, largely reproducing the spike-in scheme. PCR-based genotyping was, in dependence of the underlying PCR-assay performance, able to predict the relative amount of variant specific substitutions even in samples with low spike-in amount. The identification of variants by HTS, however, required >100 copies/ml wastewater and had limited predictive value when analyzing at a genome coverage below 60 %.
This interlaboratory test demonstrates that despite highly heterogeneous isolation and analysis procedures, overall SARS-CoV-2 GCE and mutations were determined accurately. Hence, decentralized SARS-CoV-2 wastewater monitoring is feasible to generate comparable analysis results. However, since not all assays detected the correct variant, prior evaluation of PCR and sequencing workflows as well as sustained quality control such as interlaboratory comparisons are mandatory for correct variant detection.
New paper from Wilhelm et al., 2023